Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution
[Publisher Link] [Local Copy]
Ruixuan Gao*, Shoh M. Asano*, Srigokul Upadhyayula*, Igor Pisarev, Daniel E. Milkie, Tsung-Li Liu, Ved Singh, Austin Graves, Grace H. Huynh, Yongxin Zhao, John Bogovic, Jennifer Colonell, Carolyn M. Ott, Christopher Zugates, Susan Tappan, Alfredo Rodriguez, Kishore R. Mosaliganti, Shu-Hsien Sheu, H. Amalia Pasolli, Song Pang, C. Shan Xu, Sean G. Megason, Harald Hess, Jennifer Lippincott-Schwartz, Adam Hantman, Gerald M. Rubin, Tom Kirchhausen, Stephan Saalfeld, Yoshinori Aso, Edward S. Boyden**, Eric Betzig** (2019) Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution, Science 363(6424):eaau8302. (*, equal contribution, **, co-corresponding)
INTRODUCTION
Neural circuits across the brain are composed of structures spanning seven orders of magnitude in size that are assembled from thousands of distinct protein types. Electron microscopy has imaged densely labeled brain tissue at nanometer-level resolution over near-millimeter-level dimensions but lacks the contrast to distinguish specific proteins and the speed to readily image multiple specimens. Conversely, confocal fluorescence microscopy offers molecular contrast but has insufficient resolution for dense neural tracing or the precise localization of specific molecular players within submicrometer-sized structures. Last, superresolution fluorescence microscopy bleaches fluorophores too quickly for large-volume imaging and also lacks the speed for effective brain-wide or cortex-wide imaging of multiple specimens.
RATIONALE
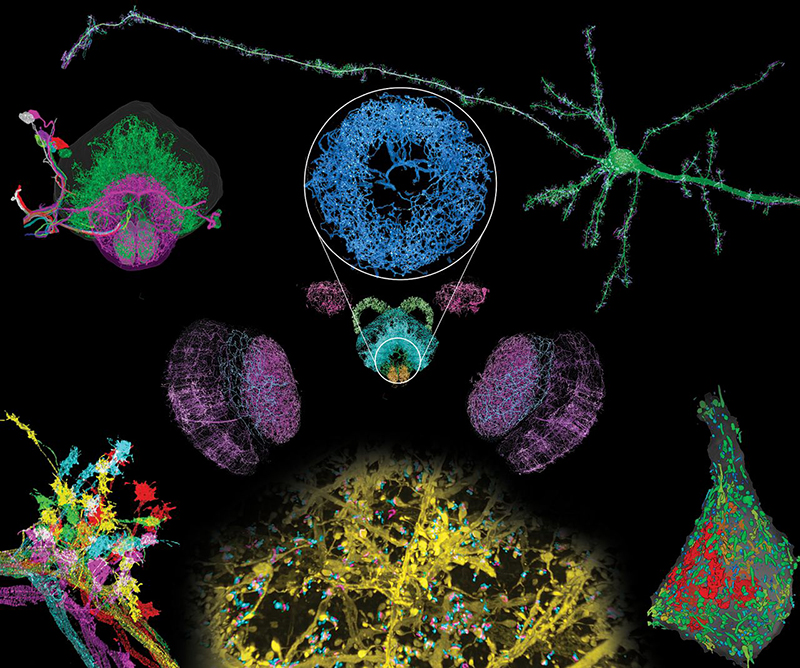
We combined two imaging technologies to address these issues. Expansion microscopy (ExM) creates an expanded, optically clear phantom of a fluorescent specimen that retains its original relative distribution of fluorescent tags. Lattice light-sheet microscopy (LLSM) then images this phantom in three dimensions with minimal photobleaching at speeds sufficient to image the entire Drosophila brain or across the width of the mouse cortex in ∼2 to 3 days, with multiple markers at an effective resolution of ∼60 by 60 by 90 nm for 4× expansion.
RESULTS
We applied expansion/LLSM (ExLLSM) to study a variety of subcellular structures in the brain. In the mouse cortex, we quantified the volume of organelles, measured morphological parameters of ~1500 dendritic spines, determined the variation of distances between pre- and postsynaptic proteins, observed large differences in postsynaptic expression at adjacent pyramidal neurons, and studied both the azimuthal asymmetry and layer-specific longitudinal variation of axonal myelination. In Drosophila, we traced the axonal branches of olfactory projection neurons across one hemisphere and studied the stereotypy of their boutons at the calyx and lateral horn across five animals. We also imaged all dopaminergic neurons (DANs) across the brain of another specimen, visualized DAN morphologies in all major brain regions, and traced a cluster of eight DANs to their termini to determine their respective cell types. In the same specimen, we also determined the number of presynaptic active zones (AZs) across the brain and the local density of all AZs and DAN-associated AZs in each brain region.
CONCLUSION
With its high speed, nanometric resolution, and ability to leverage genetically targeted, cell type–specific, and protein-specific fluorescence labeling, ExLLSM fills a valuable niche between the high throughput of conventional optical pipelines of neural anatomy and the ultrahigh resolution of corresponding EM pipelines. Assuming the development of fully validated, brain-wide isotropic expansion at 10× or beyond and sufficiently dense labeling, ExLLSM may enable brainwide comparisons of even densely innervated neural circuits across multiple specimens with protein-specific contrast at 25-nm resolution or better.
Resources associated with this Publication:
[Expansion microscopy: physical magnification with nanoscale precision]