ExCel: Super-resolution imaging of C. elegans with expansion microscopy
[Publisher Link] [Local Copy]
Chih-Chieh (Jay) Yu*, Danielle M. Orozco Cosio*, Edward S. Boyden (2020) ExCel: Super-resolution imaging of C. elegans with expansion microscopy, Methods in Molecular Biology, accepted. (*, equal contribution)
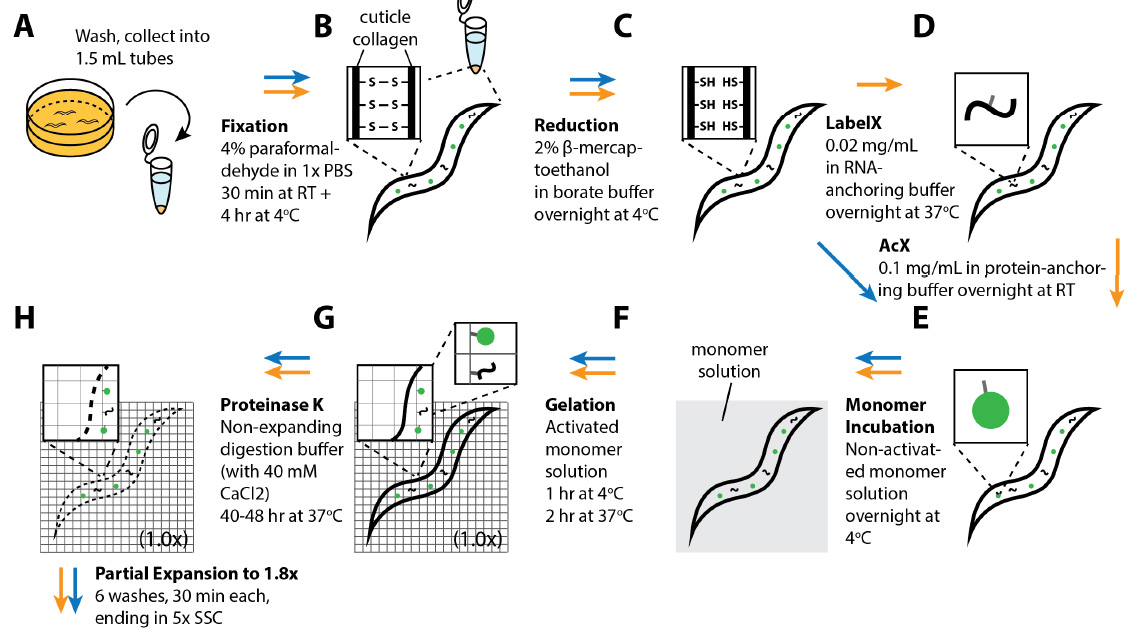
Studies of C. elegans will benefit from a powerful method for super-resolution imaging of proteins and mRNAs at any 3-D locations throughout the entire animal. Conventional methods of super-resolution imaging in C. elegans, such as STORM, PALM, SR-SIM and STED, are limited by imaging depths that are insufficient to map the entire depth of adult worms, and involve hardware that may not be accessible to all labs. We recently developed expansion of C. elegans (ExCel), a method for physically magnifying fixed whole animals of C. elegans with high isotropy, which provides effective resolutions finer than the diffraction limit, across the entire animal, on conventional confocal microscopes. In this chapter, we present a family of three detailed ExCel protocols. The standard ExCel protocol features simultaneous readout of diverse molecules (fluorescent proteins, RNA, DNA, general anatomy), all at ~70 nm resolution (~3.5x linear expansion). The epitope-preserving ExCel protocol enables imaging of endogenous proteins with off-the-shelf antibodies, at a ~100 nm resolution (~2.8x linear expansion). The iterative ExCel protocol allows readout of fluorescent proteins at ~25 nm resolution (~20x linear expansion). The protocols described here comprise a versatile toolbox for super-resolution imaging of C. elegans.
Resources associated with this Publication:
[Expansion microscopy: physical magnification with nanoscale precision]